TOP > Research > Cell Design Research Core
|
|||
Designing and controlling cellular function
By observing how they respond to small perturbations, we are designing artificial systems that act like biological ones to understand how complex and dynamical networks function within systems. Using experimental and theoretical data, we are building large-scale models for the study of molecular complexes, and gene and cellular networks. Of particular interest will be the understanding of how various spatial and temporal properties like polarity and periodicity emerge from these networks. Along with identifying the operating principles used to determine the cell state, we aim to manipulate a cell’s response into a desired effect.
|
|||
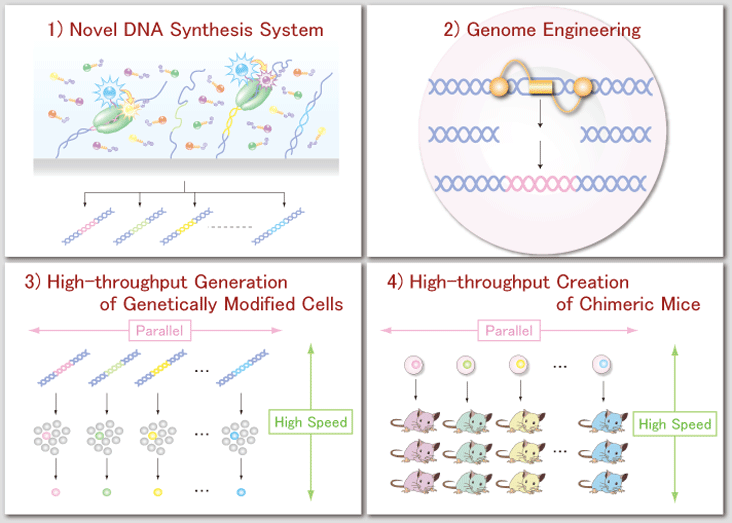
From information to a cellTo control the cell response will require bringing together a wide range of research fields for innovative technologies that include 1) the ability to supply targeted genes and proteins in a rapid and parallel manner, 2) genome engineering that can incorporate the designed and synthesized genes into the intracellular genome, and 3) high-throughput selection of the genetically modified cells and comprehensive evaluation of their function at the single cell level. To achieve these technologies, it is essential to merge advanced molecular biology, organic chemistry, and nano fabrication techniques, along with designing new equipment that can measure superior detail. Ultimately, we aim to apply these technologies to mammalian cells, especially embryonic stem cells, for organism engineering. |
|||
From cells to organismsLike the above aims, to reveal the role of cellular networks inside an organism will also require an interdisciplinary effort. Many of the technologies developed for understanding how gene and protein networks lead to cellular function will be expanded to organism engineering with the aim of characterizing the causal relationships between cellular networks and the resulting organism function. One major difference will be the addition of a fourth technology (4), a system that can generate living organisms from genetically modified cells in a rapid and parallel manner that also has high-throughput capacity for phenotypic analysis. Therefore, we aim to develop new methodologies for controlling and designing organism functions based on genomic information. |
|||
Fundamental technologies for synthetic biology and organism engineering1) Novel DNA synthesis system To artificially design and construct systems derived from complex networks of genes, proteins, and their dynamic interactions, it is essential to develop a DNA synthesis system that is fast and accurate, but also has high throughput and low cost. We therefore aim to establish a novel DNA synthesis system by merging technologies from various fields including organic chemistry, artificial protein engineering, single molecule imaging, and microfluidic microarray systems. |
|||
2) Genome engineering by artificial restriction enzymes To verify the functions of designed molecules or molecular networks in cellulo or in vivo, it is important to develop genome engineering techniques that can replace a wild-type gene with a modified one. To do this, we are developing artificially designed restriction enzymes that are composed of genome recognition and DNA cleavable units. By taking advantage of the enzymes’ ability to conduct target-specific genome scission and of a cell’s endogenous DNA repair machinery, we can arbitrary introduce a desired mutation into the unique target sequence of the intracellular genome. |
|||
3) Rapid and parallel generation of genetically modified cells combined with measuring single-cell function To control and alter the function of organisms, we will develop a high-throughput system that can generate genetically modified mammalian cells, including embryonic stem cells, by combining single-molecule fluorescence imaging technology which can detect intracellular genomic mutations with glass-based micro and nano fluidic devices. Furthermore, to evaluate the effects these artificial gene/protein networks have on cellular function, we will also develop a novel analytical system that can measure network dynamics at the single cell level. One example of the type of network this technology could measure is the expression dynamics of transcriptional regulators and their transcriptional activity. |
|||
4) A High-throughput platform for the creation of genetically modified mice The final challenge for organism engineering is to use the high-throughput technologies described above on genetically modified ES cells to conduct a comprehensive phenotypic analysis of chimeric mice. This will help establish the interplay between organismal function and cellular networks. Our goal is to manufacture a highly efficient system that produces chimeric mice consisting entirely of genetically modified ES cells. Complimenting this will be 3D-fluorescence microscopy that can characterize the cell groups and cell populations relevant to the organismal function of interest. |
|||
 |

