TOP > Research > Cell Dynamics Research Core
|
||||
|
||||
|
Japan is a world leader in the imaging of single biological molecules, having visualized a number of dynamic events like membrane receptor binding and molecular motors in solution. The next challenge is to spatially and temporally observe how such events function inside their natural environment. For this purpose we aim to develop the innovations in lasers, microscopy, and probe technology including:
These imaging techniques are expected to also allow for the quantitative analysis of a number of intracellular events in response to a stimulus including gene expression, cell polarity and membrane potential, and immunological responses. |
||||
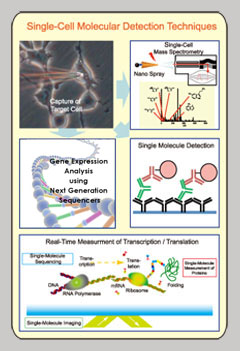
When combined with single cell manipulation techniques,innovative detection techniques can be used to analyze how different stimuli affect various cellular components. Examples of these new detection techniques include, nanospray mass spectroscopy that can be used on single cells to reveal the synthesis of various cellular components like metabolites over a given time frame. ; high-sensitivity antibodies for the purpose of detecting specific proteins inside a cell; and high-speed sequencer technology that can quantify gene expression at the single cell level. Furthermore, using these detection techniques with the aforementioned single molecule imaging, we expect to simultaneously observe intracellular changes and dynamics that will provide a more comprehensive description of cellular activity. Additionally, these techniques will also be able to observe transcription and translation in real-time to detect key mRNA base sequences and protein-protein interactions that occur during given cellular functions. |
  |
|||
Perhaps no research field has the therapeutic potential that cell stems have. However, to realize this potential, it must understand the mechanisms that make these cells so versatile and therefore applicable to a wide range of disease. A stem cell's fate comes not only from its unique features, but also how it interacts with its environment. Like all cells, stem cells make up part of a complicated and dynamic network in which each element – the cell itself or components of the cell – is in constant flux. We believe that full clinical application of stem cells can only be realized if we can control and manipulate these complex networks. By combining our new detection techniques with others methods like single molecule imaging, we expect to measure the dynamics of individual cells within large multi-cellular networks like an organ. Our goal is to reduce a cell to its most basic scale by detecting cells at previously undetectable depths inside the organ and applying observing intracellular gene and protein expressions using 3+1 spatial-temporal dimensions. Not only will such studies clarify the operating principles that drive these dynamics, they will also be the breakthroughs that lead to new life science fields. |
 |
|||

