Research
Optical microscopy is indispensable for medical and life sciences, offering
a detailed observation of living specimens in real time. We have been developing
the new technologies to quantitatively measure the behaviors of individual
elements based primarily on optical microscopy. When light is illuminated
to or emitted from a biomaterial, the light is modified and/or modulated
by the biomaterial. Measurements of this light provide information about
the biomaterial.
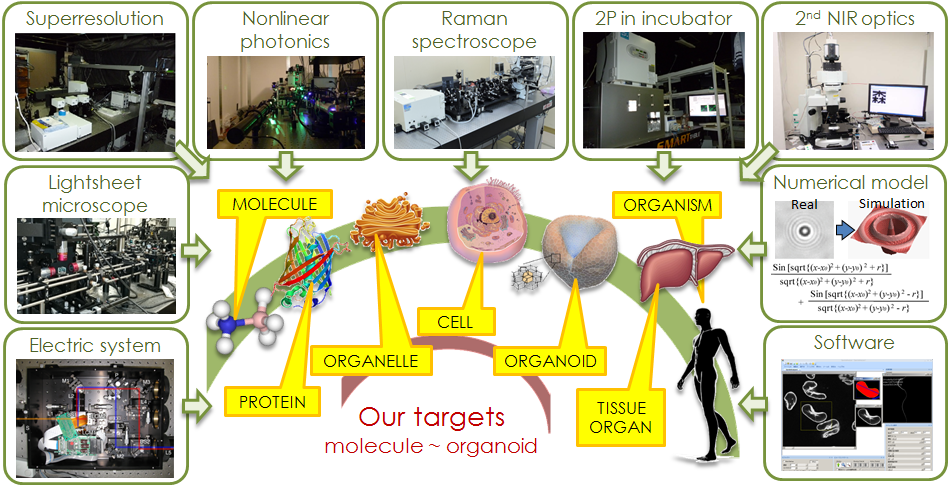

Our laboratory has developed measurement technologies to extract such biological information from the fluorescence or scattering lights. The developed techniques will help understand the fundamental mechanisms that give rise to these disparate and complex systems, thus providing significant insight on the regulation of tissues, immunity, metabolism and aging. In turn, the information will significantly improve our ability to diagnose and treat diseases and accelerate the design of new medicines.

We are now working on investigating hierarchical structure in biology. The elements composing a single-cell behave stochastically, interacting with each other with a restrained role with a certain level of freedom at the individual element level, which results in heterogeneity. Hence the cell can be considered as a complex system with a dynamic equilibrium that fluctuates stochastically between multiple states. Analogously, multiple cells interact to form even more complex systems, creating a fractal hierarchy of sorts. Both perspectives can be integrated within a common model, in which there exists transition of complex interactions between elements, be it genes, proteins or cells. Using such a model, we aim to describe the common mechanisms of these hierarchical systems.
Our developed technologies and recent biological researches are selectively
listed below.
Raman cellular Fingerprinting
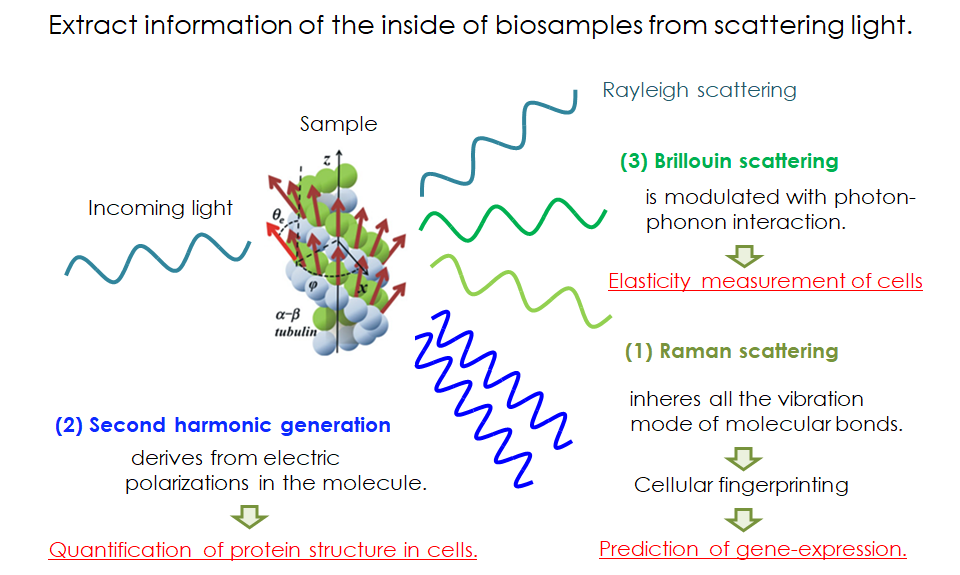
- Relationship of Raman scattering and intracellular state
- When a light illuminates to a molecule, the light is given the energy
from the molecular vibration or takes the energy to the molecular vibration,
and then scattered. This is Raman scattering and anti-stokes Raman scattering.
Therefore, the spectra peak of the Raman scattering is a little shifted
from that of the incident light, depending on the species of molecular
vibration. Molecule is composed of a lot of kinds of molecular vibrations,
like NH bond, NC double bond, benzene ring, and so on. The spectrum shape
of the molecule is measured as a convolution of those molecular vibrations.
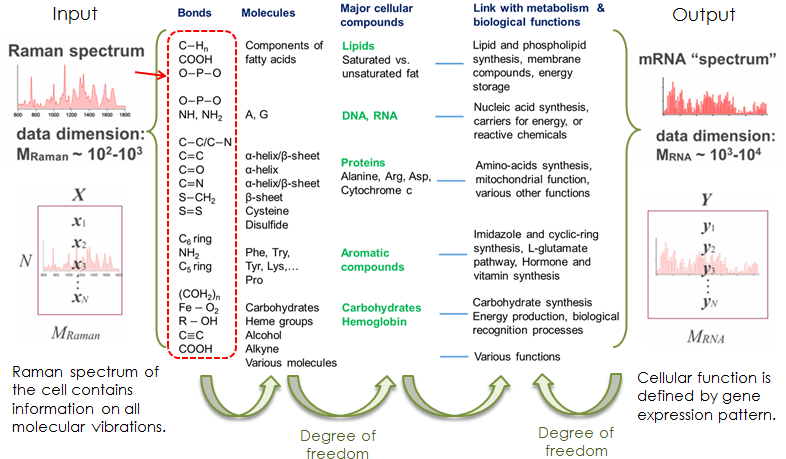
Here, let’s think about the relationship between Raman spectrum and gene-expression
of a cell. Raman spectrum depends on the molecular vibrations in a material.
These molecular bonds composes molecules in the cell. The population of
the molecules in the cell is determined by the cellular compound like lipids,
DNA, proteins and somethings. These compounds are produced depending on
the function of the cell. And, the cellular function is usually defined
by gene expression pattern. In short, the Raman spectrum possibly correlate
to the gene expression pattern via cellular functions.

- Identification of cell states/types by utilizing Raman spectrum
- While states of cells are usually defined by protein/gene expression patterns we have proposed to measure a cell’s fingerprint using its Raman spectrum as an alternative for identifying the cell state, and have found that Raman spectra can distinguish the type/states of cells. We are now applying the Raman cellular fingerprinting for medical applications, especially, evaluation of iduced pluripotent stem cells (iPS cells). We are now connecting the correlation between Raman spectra and RNA sequence in iPS cell strainsr. Those data can lead to finding the bio marker to indicate the cell state in Raman spectrum peaks.
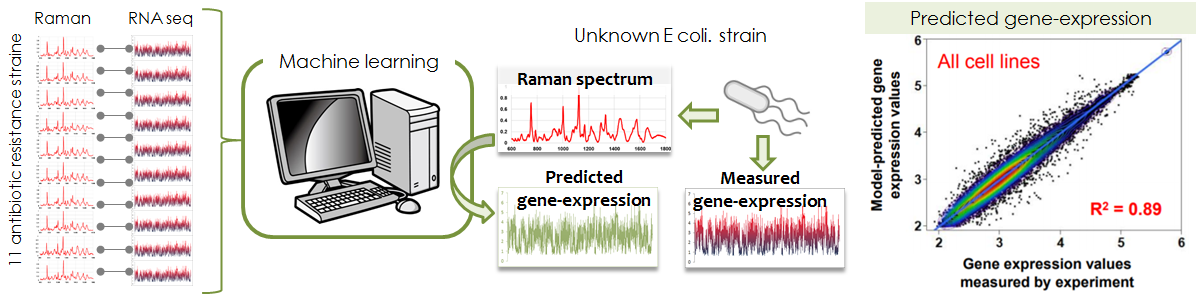
- Prediction of gene expression from Raman spectrum
- We succeeded in predicting gene-expression of antibiotic resistant bacteria
in combination with machine learning technology. Furthermore, the cluster
of the gene-Raman correlation can visualize the relationship of metabolism
and function defined by gene expression, because Raman spectrum is strongly
reflective of metabolites. The most important thing is that Raman measurement
is non-invasive unlike the current biochemical methods.

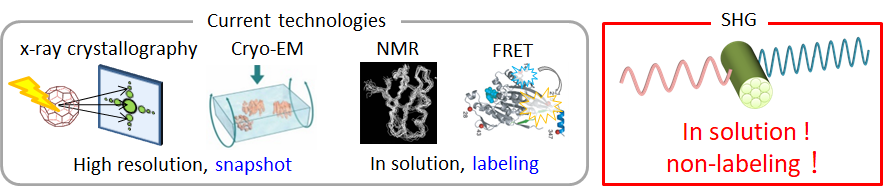
Structure analysis by utilizing Second harmonic Generation (SHG)
- Relationship of Second harmonic generation (SHG) and protein structure
- Second-harmonic generation (SHG) microscopy has been a powerful modality
to visualize fiber structures, such as fibrillar collagen in tendon, myosin
thick filaments in myosarcoma, and densely bundled microtubules in living
specimens. The SHG is a nonlinear coherent process in which new photons
with twice the energy of the incident light are generated when illuminating
a nonlinear material. The SHG susceptibility of a material is expressed
with 3rd-rank tensor (SHG tensor) originating from the structures of molecules
with permanent electric dipole moments and their alignments in macromolecular
assemblies. The SHG signal, therefore, potentially carries the structural
information of protein, which can be read out optically without labeling.
We propose adding SHG polarization measurements as another modality in
the pallet of structural analysis methodologies.

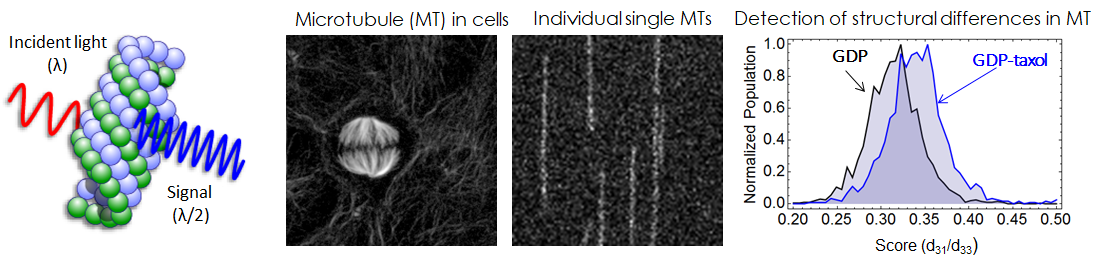
- Optical detection of structural changes within a protein
- A major reason why SHG has not been applied for structural analysis is its extreme low signal-to-noise (S/N) ratio to detect structural differences intra protein. First of all, we newly constructed a transmittion-type SHG microscope combined wiht a fast polarization controllable device in order to measure the SHG tensor. Our SHG microscopy finally achieved high sensitivity to detect weak SHG signal emmited from an indivisual single microtuble fiber. And we could perform polarization dependence measurement for the observation of structural dynamics of single microtubules without labeling.

- Medical application based on SHG microscopy
- We are going to establish quality evaluation of artificial cardiac cells derived from induced pluripotent cells (iPS cells) based on SHG microcscopy. Actomyosin activity can be evaluated with SHG signal because structure of actomyosin dramatically changes during force generation. We established a method to evaluate actomyosin activity in cardiac cells derived from iPS cells.
Mechanism of pluripotency/difference in embryonic stem cells (ES cells)
- Investigation of the cellular communication during early ES differentiation
- Embryonic stem cells (ES cells), which can differentiate into all types of cells, have been suggested to differentiate in a collective manner despite the absence of leading cells or a circadian clock. The differentiation pattern of ES cells varies within each individual colony. We quantitatively measured the expression of Nanog and Oct3/4 of single cells in individual colonies every day for 8 days after the induction of cell differentiation. It was found that the neighboring cells communicate with each other and that the communication manner that control the degree of the expression fluctuation changes between the early and later stage of differentiation. In short, Two criteria for the avalanche of the collective-state transition were revealed: spontaneous fluctuation and cell-cell cooperativity.
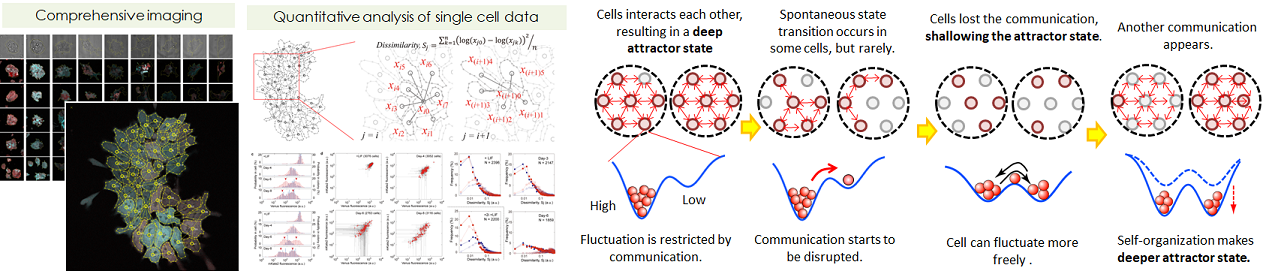

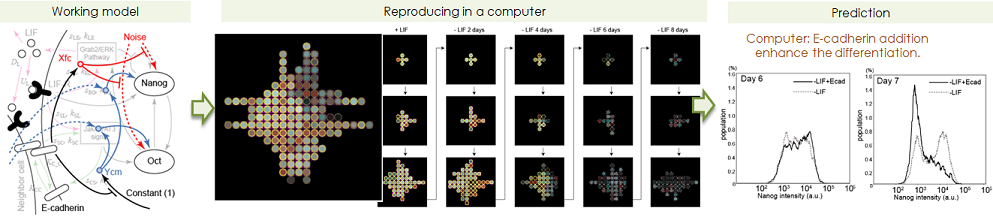
- Prediction of differentiating behaviour on computer, and prediction of perturbation.
- We constructed numerical toy models how cell-cell communication impact
the colonial heterogeneity formation, based on quantitative measurement
data. To support the essentiality of cell-cell communication by cell contact,
we had a strategy as following explanation. First we constructed multiple
scenarios. Afterwards, to confirm the validity of them by prediction the
actual wet experiment. This model gave us the idea that a cell can sense
the Nanog/Oct4 expression levels of the surrounding cells. The expression
pattern of a cell-cell adhesion molecule, E-cadherin is similar to the
tightness of cell-cell communications, therefore E-cadherin is appropriate
to link to the simulation gene network pathway. The model prected that
the extra addition of E-cadherin plays a role not only inhibiting gating
to differentiation but also enhancing the differentiation, and this is
actually happend in real experiment.

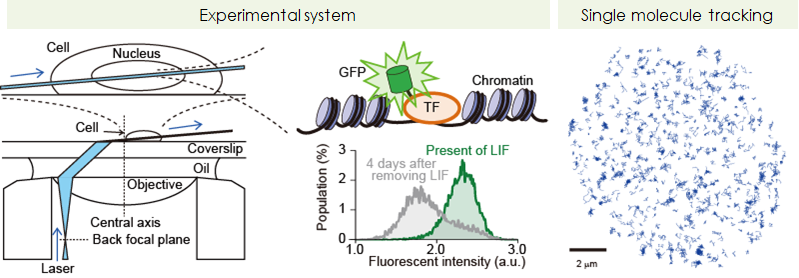
- Biophysical revealing of functions of core-transcription factors, Nanog and Oct4
- In order to reveal the mechanism of pluripotency maintenance from the biophysical aspect, we quantitively and comprehensively measured interactions of single molecules of Nanog and Oct4 on the target DNA in living ES cells. By analyzing more than 2,000,000 molecules in 1000 cells, we found the vairous biological finding that no one have discussed.