|
Systems Biology and the history of LSB
Systems Biology is a natural extension of molecular biology, and can be defined as "biology after the identification of key genes". We envision systems-biological research as a multistage process, beginning with the comprehensive identification and quantitative measurement of individual system components and their networked interactions, leading to the ability to control existing systems toward a desired state and to design new systems based on an understanding of structure and underlying principles. When the term Systems Biology was coined and this type of integrative research was advocated beginning around the year 2000, a number of interesting concepts had already been introduced. At that time, few systems-biological approaches were well-developed and their application to fundamental biological questions was limited.
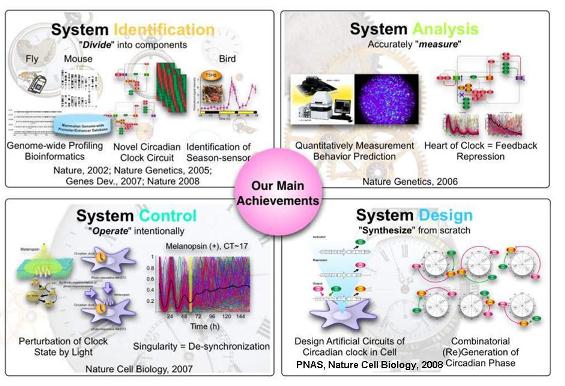
Figure 1. Strategies and technologies for systems-biological research and their application to specific questions regarding the biological clock. Systems-biological research starts with comprehensive identification (top left). In this step, individual system components and their network interactions are comprehensively identified. In the second step, to derive the design principles of the target system, the behavior of the system is predicted and validated through perturbation and accurate measurements (top right). An understanding of the design principles of the system is essential to derive a method of controlling the system to achieve desired behavior (bottom left). Finally, the level of understanding is confirmed by reconstruction of the system (bottom right).
From 2001 to 2010 we worked to establish experimental systems biology at the molecular level, investigating system-level properties of complex and dynamic biological systems such as the mammalian circadian clock. We have applied these systems-biological approaches to specific questions (Fig. 1), leading to the identification of clock circuits in mammals and flies and a spring hormone in birds (Ueda H.R. et al., Nature 2002, Ueda H.R. et al., Nat Genet 2005, Matsumoto A., Ukai-Tadenuma M., Yamada R.G. et al., Genes Dev 2007, Nakao N. et al., Nature 2008); uncovering a pivotal role for feedback repression in mammalian circadian clocks (Sato T.K., Yamada R.G., Ukai H. et al., Nat Genet 2006); discovering desynchronization as the underlying mechanism for singularity behaviors in mammalian circadian clocks (Ukai H., Kobayashi T.J. et al., Nat Cell Biol 2007); and developing the design principles of clock-controlled elements and circadian transcriptional outputs (Kumaki Y. et al., PNAS 2008, Ukai-Tadenuma M., Kasukawa T. et al., Nat Cell Biol 2008). Based on these previous achievements, we strongly feel that it is now time to take the next step forward toward experimental systems biology at the organism level.
Research Aim
We aim to realize experimental systems biology at the organism level. This aim has been extremely difficult so far and cannot be realized without focused efforts toward multi-scale understanding of biological systems. To this end, we selected mammalian circadian clocks as a model system, and will implement, in the first five years, multi-scale research activities on this system, covering a range of scales: (i) molecule-to-cell, (ii) cell-to-tissue, and (iii) tissue-to-organism. For each layer, we will solve at least one fundamental question regarding biological time by integrating multi-disciplinary technologies.
Specific Foci
(i) At the molecule-to-cell scale, we will elucidate the robustness and flexibility of transcriptional networks. In particular, we will focus on robustness against noise and epigenetic flexibility in the circadian clock.
(ii) At the cell-to-tissue scale, we will understand the collective behavior of heterogeneous cells. Our major area of focus will be synchronization and coupling- dependent oscillations in the central clock tissue.
(iii) At the tissue-to-organism scale, we will reveal timekeeping mechanisms performed by multi-tissue (or multi-brain-region) systems. We will specifically focus on the cellular circuits controlling the sleep/wake cycle and address the hourglass mechanism of sleep, a homeostatic- and circadian-dependent regulation of sleep amount and timing. To approach such complicated cellular circuits, we are currently developing next-generation mouse genetics, where we can produce genetically modified mice in a high-throughput fashion (Fig. 2).
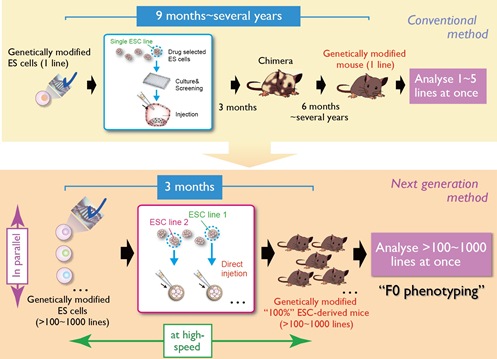
Figure 2. Scheme of next-generation mouse genetics.
In the conventional method for production of genetically-modified mice, a single line of gene-targeted ES cells is injected into host embryos (typically blastocysts) to generate chimera mice comprising a mixture of ES- and host-derived cells. In addition, multiple mating procedures are needed to generate the desired genetically modified mouse strain, which typically takes from 9 months to several years (top). To develop a high-throughput method of mouse genetics (next-generation mouse genetics), we are trying to 1) shorten the process by realizing F0 phenotyping with ~100% ES cell-derived mice (the same generation at which chimeras are typically produced) to skip the mating procedures, and 2) parallelize the process by simultaneous preparation of multiple lines of ES cell clones and their direct transfer to host embryos (single colony injection). We aim to use this novel high-throughput method to produce hundreds to thousands of strains of genetically modified ~100% ES cell-derived mice (bottom).
|
| Page Top |