|
IV. Design of Clocks
The next step is System Design, reconstruction of the system using hypothesized design principle that has been revealed through the efforts of combination strategy of system identification, system analysis, and system control for validating the sufficiency of the system components.?
Three specific features of circadian oscillatory wave are the period length, the amplitude, and the peak phase. We treat latter two in this section. At first we challenged to reach the amplitude synthesizing mechanism through design of artificial clock-controlled elements (Section 3-IV-1). To the next, we tried to reveal the phase determination principle using our in vitro cycling assay system as "physical simulator" (Section 3-IV-2).
IV-1. Design of the Clock-controlled Elements: Amplitude
Mammalian circadian clock consists of complexly integrated regulatory loops mediated via three clock-controlled elements (CCEs, Section 3-I-1), E-box, D-box and RRE. Circadian oscillatory information are transmitted through these CCEs to the expression of downstream genes (clock controlled genes, CCGs) and at last reflected to physiological and metabolic processes. To transmit information to the downstream, it is important to maintain important features of waves, such as amplitude. However, we still did not know how the transcriptional regulatory system maintains the high-amplitude oscillation. As an initial step toward an understanding of a dynamic transcriptional regulatory system, we designed a synthetic promoter based on our previous analysis descrived above (Section 3-I-2), which enabled the identification of the direct targets of mammalian circadian clocks with hidden Markov models (HMMs, Section 3-I-2) of CCEs.
To investigate the characteristics of CCEs, we emitted from our HMMs synthetic regulatory elements that do not exist in nature. If our HMMs represented the functional response elements?to natural transcription factors, then synthetic regulatory elements derived from these models would mediate rhythmic transcription as well. We emitted millions of sequences from the E-box, D-box and RRE models, and filtered out those that exist naturally. Furthermore, in an attempt to not unduly focus our attention on outliers we required that all candidates adhere to the consensus rules for each CCEs. For the remaining sequences, we chose the highest and lowest scoring synthetic representatives ("high-scoring" and "low-scoring", Fig. 3-IV-1-1 A). To test these elements, we constructed a synthetic promoter (three tandem repeats of these elements fused to the SV40 promoter) driving a dLuc. These constructs were transfected into NIH3T3 cells and we subsequently measured their transcriptional activity (Fig. 3-IV-1-1 B). All high-scoring elements showed high-amplitude circadian transcriptional activity equivalent to known elements from canonical clock genes (E-box of Per1, D-box of Per3 and RRE of Bmal1 are used as 1.0, Fig. 3-IV-1-1 C). In contrast, the low-scoring elements showed relatively low-amplitude transcriptional activity, despite the presence of consensus CCEs core sequences (Fig. 3-IV-1-1 C). These results reveal that the amplitude information is encoded in sequences adjacent to the consensus element, and shows the utility of this comparative genomics approach in carrying out the synthetic design of dynamic cis-acting elements.
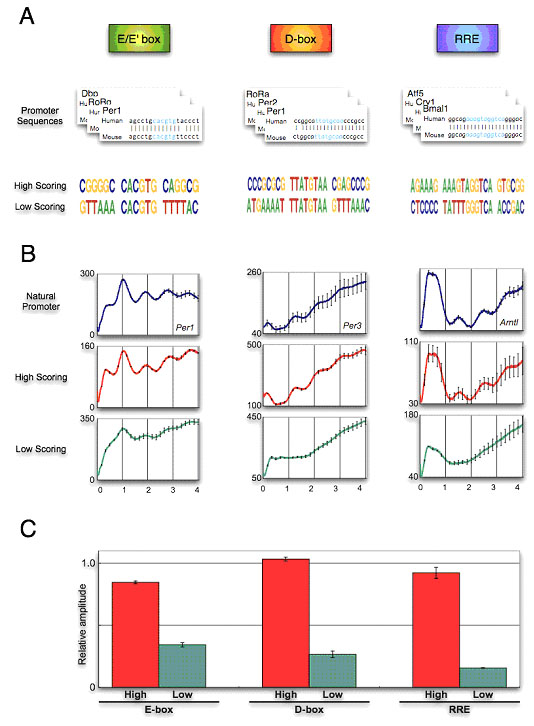
Figure. 3-IV-1-1 Computational design and experimental realization of high-amplitude circadian transcriptional activity. (A) HMM-based design of the high- and low-scoring elements. (B) Bioluminescence from synthetic elements inserted into SV40-dLuc in NIH3T3 cells. SV40-dLuc containing known CCEs and SV40-dLuc with no insert are act as the positive and negative controls. The negative control of the D-box element was also used for the RRE as experiments for these elements were performed at the same time. (C) Amplitude of bioluminescence activity driven by synthetic elements relative to that of positive controls. The relative amplitude of positive controls is 1.0.
We further hypothesized that this adjacent DNA influenced the DNA-binding affinity of the regulatory factors leading to altered retention and, as a consequence, altered amplitude. To test this notion, we analyzed the affinity of activators and repressors to these circadian response elements using competitive DNA binding assays (Fig. 3-IV-1-2 A). For the D-box and RRE elements, high-scoring elements showed approximately the same affinity as that of positive controls for both canonical direct activators and repressors, while low-scoring elements of D-box and RRE showed relatively weak affinity, thereby confirming the hypothesis.
Surprisingly, in the case of E-box low-scoring sequences had higher affinity to Bmal1/Clock activator complex (4.8 times higher than positive control) than high-scoring sequences or the positive control, whereas the low-scoring E-box sequence showed approximately the same affinity to the DEC1 repressor. To assist in interpreting these results, we adopted use of in silico modeling and examined as parameters the affinity for activators and repressors and amplitude as the output of the model. Interestingly, this analysis showed that a high affinity activator complex coupled with a normal affinity repressor complex capitulated lower amplitude rhythms (Fig. 3-IV-1-2 B), suggesting that the enhanced retention of an activator alone can lead to saturation on a promoter and consequently dampen amplitude. Further in silico analysis revealed not only the affinity strength of the activator and repressor but also the appropriate balance of affinity between activators and repressors?are necessary for high-amplitude circadian oscillation.
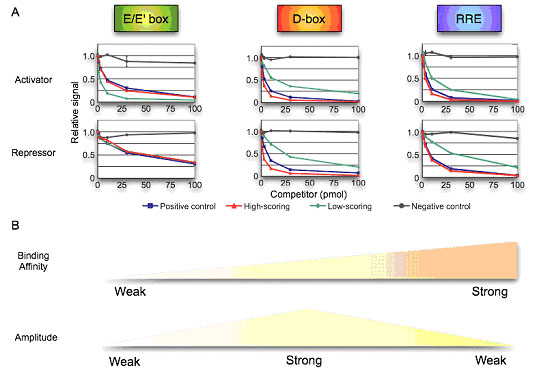
Figure. 3-IV-1-2 Defining the relationship between affinity and amplitude. (A) Binding affinity between synthetic elements and DNA binding activators or repressors was detected by competitive DNA binding assays. The binding between labeled oligonucleotides of positive control elements (10 pmol) and regulators were competed by each of the unlabeled oligonucleotides (0, 1, 3, 10, 30 and 100 pmol) for positive control (blue), high-scoring (red), low-scoring (green) or negative control elements (black). Known and mutated CCEs act as the positive and negative controls. Bmal1/Clock, Dbp, Rorα were used as DNA binding activator, and Dec1, E4BP4, RevErbAα were used as DNA binding repressors of E-box, D-box and RRE. The relative signal without competitor is 1.0. (B) Balance of binding affinity and amplitude. The enhanced retention of an activator alone can lead to saturation on a promoter and consequently dampen amplitude.
We have applied a synthetic?strategy to the understanding of a dynamic transcriptional regulatory system, the mammalian circadian clock. We have taken these in silico models and designed synthetic elements that exhibit high-amplitude transcriptional rhythmicity as efficiently as naturally occurring canonical regulatory elements. Biochemical experiments on these synthetic elements revealed the importance of DNA affinity balance between activators and repressors of a particular element in generating high-amplitude circadian transcriptional output. The synthetic approaches discussed here are especially timely as genomics tools and are increasingly being used in uncovering the complexity and flexibility of transcriptional regulatory circuits. Seen in this light, therefore, the design methods presented here will promote and enhance understanding of the biology mediated by complex and dynamic transcriptional regulation including the mammalian circadian clock.
We have submitted a manuscript regarding this study to a scientific journal.
IV-2. Design of Circadian Circuits: Phase
Although the network structure composed by clock genes and clock-controlled DNA elements (CCEs, Section 3-I-1) has been described comprehensively, the dynamic principles governing this transcriptional circuit remains elusive. One of the key issues concerning the logic of the mammalian circadian clock is how the circadian oscillating genes' expression peaks ("phases") are determined. In attempting to elucidate this issue, we have started a project for understanding the molecular logic of phase-controlling system(s). For this purpose, we have extended?our in vitro cycling assay system to the "physical simulator", with which we can implement artificial circuits of interest. We used this system to prove the sufficiency of the components we predict in natural circadian phase-controlling mechanism.
Mammalian circadian clocks are composed of complexly integrated transcriptional regulatory loops. To reach a system-level understanding of the circadian clock controlling system, we focused on the three main transcriptional regulation elements (CCEs), E/E'-box D-box and RRE (Section 3-I-1). We first focused on the day-time transcriptional regulation mediated via the D-box. The transcriptional activator DBP activates genes expression via the D-box, while the transcriptional repressor E4BP4 represses?expression (Fig. 3-IV-2-1 A, upper panel). Dbp is regulated by the E-box, the morning control element, while E4bp4 is under the regulation of the RRE, the night-time control element. On the basis of this transcriptional circuit information, we hypothesized that a morning activator and a night-time repressor can determine the day-time transcriptional output mediated through the D-box. Similarly, RRE activators (e.g. Rorα ) are expressed during the day-time under the control of the D-box, and the RRE repressors (e.g. RevErbAα ) seem strongly influenced by morning element (E'-box, Fig. 3-IV-2-1 A, lower panel). We have therefore also been able to make the simple hypothesis that a day-time activator and a morning repressor can specify the night-time transcriptional output mediated through the RRE. To test these hypotheses, we adopted a synthetic approach in order to physically simulate an identified structure, and observed the resulting dynamics using artificial transcriptional circuits.
To design and implement artificial transcriptional circuits in mouse NIH3T3 cells, which has a self-oscillating circadian clock, we developed an in vitro cycling assay system composed by the following three components; an artificial activator (a destabilized GAL4-VP16 fusion protein: dGAL4-VP16), an artificial repressor (a destabilized GAL4 protein: dGAL4), and an output reporter (a destabilized luciferase (dLuc ) gene driven by a minimal TATA box fused with four tandem repeats of UAS, the GAL4 binding sequence) (Fig. 3-IV-2-1 B). In this system, the input from artificial activator and repressor competitively regulate the output and reporter gene expression through the UAS (Fig. 3-IV-2-1 B, lower panel) and the dynamics of the output could be monitored through the light intensity of the expressed dLuc protein under a real time bioluminescence measuring system. As the artificial activator and repressor are driven by the SV40 basic promoter fused with three tandem repeats of CCEs, we could control the expression timing of these artificial regulators via the morning (E'-box), day-time (D-box) or night-time (RRE) elements. We reasoned that if (and only if) the phases of the transcriptional activator(s) and repressor(s) are acceptable determinants of the phase of the downstream transcriptional output, then, it should be possible to generate the natural phases using synthetic transcriptional regulators and promoters. If this were proved to be so, the findings would prove to be very significant, as transcription factors are regulated by various post-transcriptional mechanisms, including translation, phosphorylation, ubiquitination, sumoylation, and nuclear transportation, which are thought to contribute, at least in part, to the phases decision of the downstream transcriptional outputs.
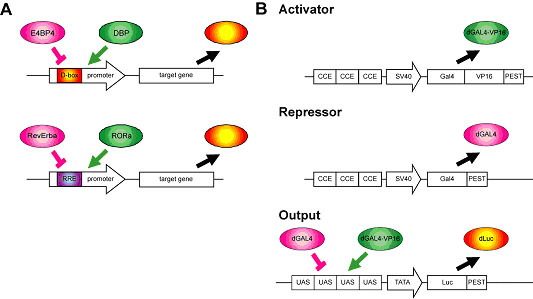
Figure. 3-IV-2-1 Synthetic transcriptional circuit of mammalian circadian clocks (A) Hypothetical models of day-time and night-time outputs regulation. In natural circuits, Dbp and E4bp4 control the expression via the D-box. Similarly, RevErbAα and Rorα participate in the regulation via the RRE. (B) The artificial transcriptional system. dGAL4-VP16 and dGAL4 were used as the activator and repressor. These transcriptional factors were expressed under the control of three tandem repeats of a CCE (E'-box of Per2, D-box of Per3 or RRE of Bmal1). The artificial activator and repressor competitively bind the four tandem repeats of UAS in the artificial promoter to regulate the output reporter gene dLuc. For observation of the dynamic behaviour of this artificial circuit, NIH3T3 cells were transiently transfected with plasmids harboring the activator, repressor or reporter, and the reporter activity was monitored under a real time bioluminescence measuring system.
Using the in vitro cycling assay system as a physical simulator, we first tested our hypothesis for day-time output by examining the dynamic behaviour of the transcriptional output generated from the competition of an artificial morning activator controlled via E'-box and night-time repressor controlled via RRE. The phases of the activator and repressor in this experiment were detected at circadian time (CT) 4.0 ± 0.29 (n=2), and CT17.1 ± 0.37 (n=2). The transcriptional output driven by these regulators exhibited circadian oscillation with a phase at CT7.7 ± 0.85 (n=2), which is very close (≤ 1.0 h) to the corresponding natural day-time (D-box) phase, CT8.7 ± 1.13 (Fig. 3-IV-2-2 A). These results suggest that morning activation and night-time repression are sufficient to determine the day-time transcriptional output. Importantly, we could not observe high-amplitude circadian oscillation by expressing the morning activator or night-time repressor alone. From these, we concluded that both morning activation and night-time repression are necessary for day-time output.
We next tested the hypothesis for night-time output by examining another artificial circuit consisting of an artificial day-time activator under D-box control and a morning repressor under E'-box control. The phases of a transcriptional activator and repressor in this experiment were at CT8.9 ± 0.28 (n=2) and CT3.9 ± 0.13 (n=2). The transcriptional output driven by the regulators exhibited circadian oscillation with its phase at CT16.6 ± 1.04 (n=2), which is very close (≤ 1.0 h) to the corresponding natural night-time phase (CT17.0 ± 0.81) (Fig. 3-IV-2-2 B). These results also suggest that day-time activation and morning repression are necessary and sufficient to generate the circadian night-time output, implying that the input phases of transcriptional regulators can determine the phases of transcriptional output. These findings led us to reason that various combinations of transcriptional regulators with CCEs for the three basic circadian phases (morning, day-time, and night-time) might not only generate the basic phases, but also other phases. Indeed, we succeeded in generating various phases though simple combinations of the transcriptional regulators of the three basic circadian phases (Fig. 3-IV-2-2 C-F).
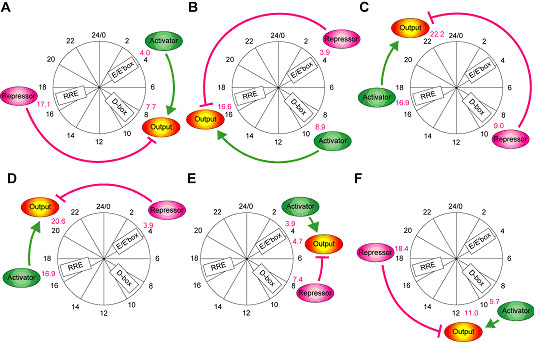
Figure. 3-IV-2-2. Promoter activities of an activator, repressor and output in different artificial transcriptional circuits. Morning activator under E'-box control and night-time repressor under RRE control (A); day-time activator under D-box control and morning repressor under E'-box control (B); night-time activator under RRE control and day-time repressor under D-box control (C); night-time activator and morning repressor (D); morning activator and day-time repressor (E); and day-time activator and night-time repressor (F). The schemes summarize the representative promoter activities of each artificial circuit monitored by bioluminescence from NIH3T3 cells, where an activator, repressor and output phases are indicated with their peak time indicated by circadian time (red numbers).
Importantly, even if the phase of the activator was not changed, the advanced or delayed phase of the repressor led to a corresponding change in the phase of transcriptional output (Fig. 3-IV-2-2). Similarly, a change in the activator phase also led to a corresponding alteration in the output phase (Fig. 3-IV-2-2). We can deduce from these findings that mammalian circadian clocks can start from three basic phases to the final-practically continuous-phases through transcriptional cascades, which have been observed in vivo in the central and peripheral clock tissues (e.g. SCN and liver). Moreover, this concept is supported by our recent observations that many transcription factors are rhythmically expressed at various phases in the liver (Section 3-V-1) and that some seem to be under the direct control of circadian clock via CCEs. In addition, our in vitro findings lead to the concept that the natural transcription activator(s) and repressor(s) which bind the similar DNA elements are expected to generate additional output phases because they are expressed in different in vivo phases.
In these experiments, we showed that the transcriptional regulation of upstream transcription factors can determine the phase of the downstream output. This study presents a synthetic approach to the "proof-by-synthesis" of transcriptional logic, which provides us with a new strategy, not only to investigate the requirements for identified components and/or their interactions but also to reveal as-yet unidentified components or interactions. These results reveal design principles for elucidating continuous transcriptional outputs observed in vivo and for the logical design of artificial promoters working at novel phases. Logical synthesis of artificial circuits with defined structure and observation of their dynamics provide an alternative strategy applicable for the investigation of complex and dynamic biological systems such as mammalian circadian clocks. These results are under submission to a scientific journal.
|
| Page Top |